02_pyscf-HF_turbo-VMC_molecules
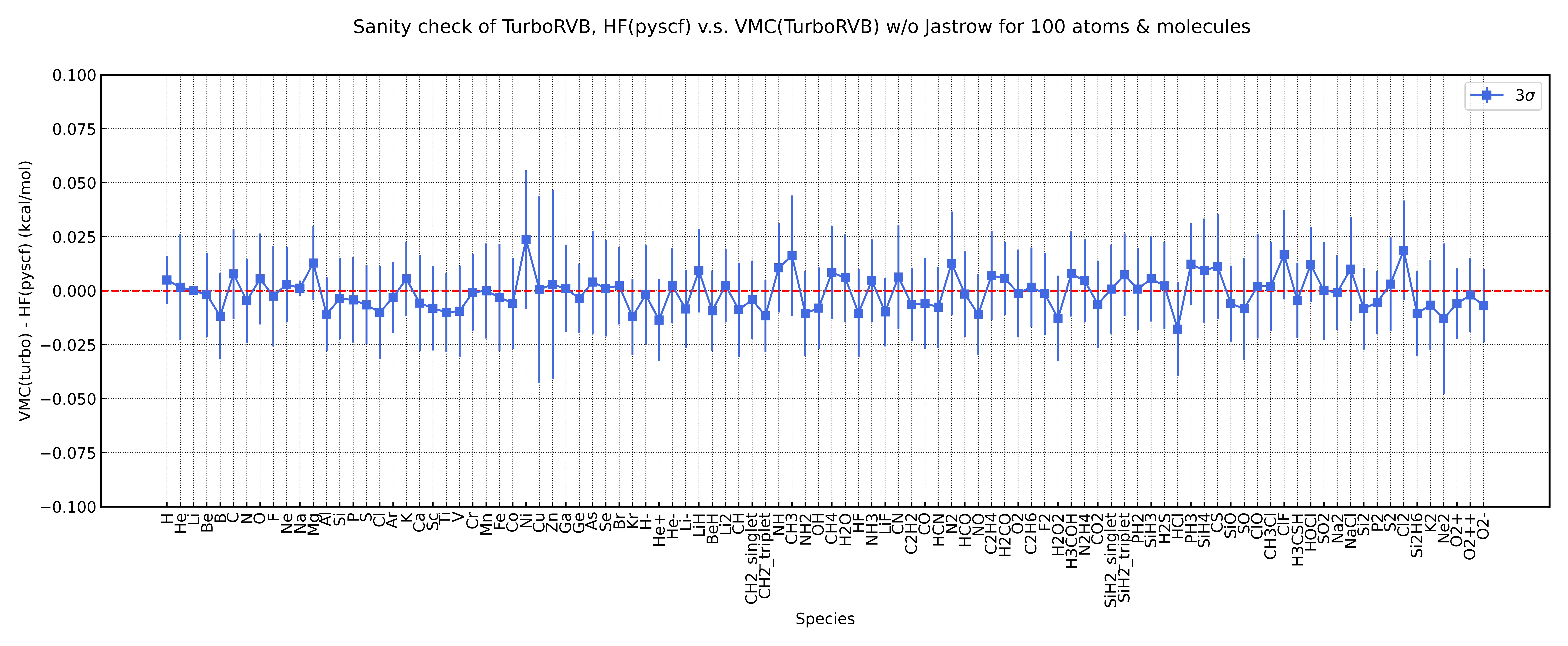
From this tutorial, you can learn how to compare HF energies obtained by pySCF and VMC energies obtained by TurboRVB for 100 molecules using turboworkflows. Here is a python script to compute it. You can download the csv and geometry files for this tutorial from here.
#!/usr/bin/env python
# coding: utf-8
# python packages
import os, sys
import numpy as np
import pandas as pd
from ase import Atoms
from ase.io import write, read
# turboworkflows packages
from turboworkflows.workflow_encapsulated import eWorkflow
from turboworkflows.workflow_lrdmc_ext import LRDMC_ext_workflow
from turboworkflows.workflow_vmc import VMC_workflow
from turboworkflows.workflow_pyscf import PySCF_workflow
from turboworkflows.workflow_trexio import TREXIO_convert_to_turboWF
from turboworkflows.workflow_vmcopt import VMCopt_workflow
from turboworkflows.workflow_lanchers import Launcher, Variable
from turboworkflows.workflow_prep import DFT_workflow
# read molecules and its info.
mol_info=pd.read_csv("data_sanity_check.csv")
mol_calc=mol_info[mol_info["Flag"]==True]
# info list:
species_list=list(mol_calc["Species"])
type_list=list(mol_calc["Type"])
label_list=list(mol_calc["Label"])
scf_newton_list=list(mol_calc["scf_newton"])
pyscf_basis_list=list(mol_calc["pyscf_basis"])
pyscf_ecp_list=list(mol_calc["pyscf_ecp"])
charge_list=list(mol_calc["Charge"])
neldiff_list=list(mol_calc["Neldiff"])
geom_ref_list=list(mol_calc["Geometry Reference"])
pid=os.getpid()
with open("turboworkflows.pid", "w") as f: f.write(str(pid)+'\n')
root_dir=os.getcwd()
result_dir=os.path.join(os.getcwd(), "results")
os.makedirs(result_dir, exist_ok=True)
os.chdir(result_dir)
cworkflows_list=[]
for species,type,label,scf_newton,pyscf_basis,pyscf_ecp,charge,neldiff,geom_ref in zip(species_list,type_list,label_list,scf_newton_list,pyscf_basis_list,pyscf_ecp_list,charge_list,neldiff_list,geom_ref_list):
mol_root_dir=os.path.join(result_dir, label)
#copy or generate xyz file.
if type=="atom":
at = Atoms(species, positions=[(0, 0, 0)])
write(f"{species}.xyz", at)
elif type=="molecule":
at = read(os.path.join(root_dir,"geometry", f"{species}.xyz"))
write(f"{species}.xyz", at)
else:
sys.exit()
pyscf_HF_workflow = eWorkflow(
label=f'pyscf-HF-workflow-{label}',
dirname=os.path.join(mol_root_dir, f'pyscf-HF-workflow'),
input_files=[f"{species}.xyz"],
workflow=PySCF_workflow(
## structure file (mandatory)
structure_file=f"{species}.xyz",
## job
server_machine_name="kagayaki",
cores=128,
openmp=128,
queue="SINGLE",
version="stable",
sleep_time=600, # sec.
jobpkl_name="job_manager",
## pyscf
pyscf_rerun=False,
pyscf_pkl_name="pyscf_genius",
charge=charge,
spin=neldiff,
basis=pyscf_basis,
ecp=pyscf_ecp,
scf_method="HF",
dft_xc="NA",
pyscf_output="out.pyscf",
pyscf_chkfile="pyscf.chk",
solver_newton=scf_newton,
twist_average=False,
exp_to_discard=0.00,
kpt=[0.0, 0.0, 0.0], # scaled_kpts!! i.e., crystal coord.
kpt_grid=[1, 1, 1]
)
)
cworkflows_list+=[pyscf_HF_workflow]
#continue #to check pyscf convergences
trexio_HF_workflow = eWorkflow(
label=f'trexio-HF-workflow-{label}',
dirname=os.path.join(mol_root_dir, f'trexio-HF-workflow'),
input_files=[Variable(label=f'pyscf-HF-workflow-{label}', vtype='file', name='trexio.hdf5')],
workflow=TREXIO_convert_to_turboWF(
trexio_filename="trexio.hdf5",
twist_average=False,
jastrow_basis_dict={},
max_occ_conv=1.0e-4,
trexio_rerun=False,
trexio_pkl_name="trexio_genius"
)
)
vmc_HF_workflow = eWorkflow(
label=f'vmc-HF-workflow-{label}',
dirname=os.path.join(mol_root_dir, f'vmc-HF-workflow'),
input_files=[Variable(label=f'trexio-HF-workflow-{label}', vtype='file', name='fort.10'),
Variable(label=f'trexio-HF-workflow-{label}', vtype='file', name='pseudo.dat')],
workflow=VMC_workflow(
## job
server_machine_name="fugaku",
cores=2304,
openmp=1,
queue="small",
version="stable",
sleep_time=9600, # sec.
jobpkl_name="job_manager",
## vmc
vmc_max_continuation=2,
vmc_pkl_name="vmc_genius",
vmc_target_error_bar=1.0e-5, # Ha
vmc_trial_steps= 150,
vmc_bin_block = 10,
vmc_warmupblocks = 5,
vmc_num_walkers = -1, # default -1 -> num of MPI process.
vmc_twist_average=False,
vmc_kpoints=[],
vmc_force_calc_flag=False,
vmc_maxtime=84600,
)
)
cworkflows_list+=[vmc_HF_workflow, trexio_HF_workflow]
vmc_HF_workflow_gpu = eWorkflow(
label=f'vmc-HF-workflow-{label}-gpu',
dirname=os.path.join(mol_root_dir, f'vmc-HF-workflow-gpu'),
input_files=[Variable(label=f'trexio-HF-workflow-{label}', vtype='file', name='fort.10'),
Variable(label=f'trexio-HF-workflow-{label}', vtype='file', name='pseudo.dat')],
workflow=VMC_workflow(
## job
server_machine_name="m100",
cores=512,
openmp=1,
queue="small",
version="stable",
sleep_time=9600, # sec.
jobpkl_name="job_manager",
## vmc
vmc_max_continuation=2,
vmc_pkl_name="vmc_genius",
vmc_target_error_bar=1.0e-5, # Ha
vmc_trial_steps= 150,
vmc_bin_block = 10,
vmc_warmupblocks = 5,
vmc_num_walkers = -1, # default -1 -> num of MPI process.
vmc_twist_average=False,
vmc_kpoints=[],
vmc_force_calc_flag=False,
vmc_maxtime=84600,
)
)
cworkflows_list+=[vmc_HF_workflow_gpu]
launcher=Launcher(cworkflows_list=cworkflows_list, dependency_graph_draw=True)
launcher.launch()